Identification of mRNA subcellular localization
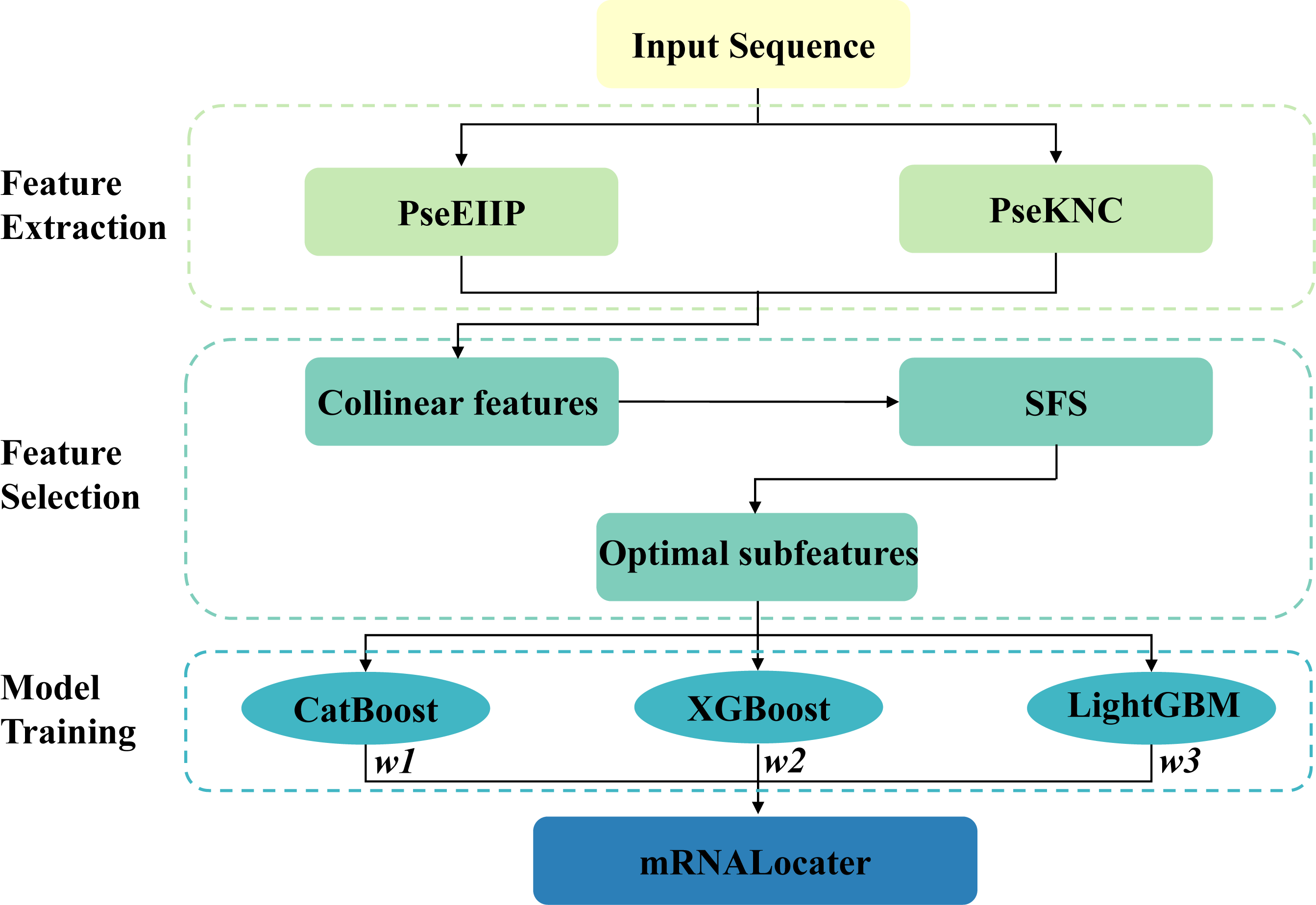
Electron-ion interaction pseudo-potential (PseEIIP) values of trinucleotides and pseudo k-tuple nucleotide composition (PseKNC) were used to transform sequences of various length into fixed-length numerical vectors. Subsequently, a two-step optimal feature selection scheme was used to find out the optimal feature subset, which were used as the input features of lightGBM, XGBoost and CatBoost to build the ensemble prediction model mRNALocater.
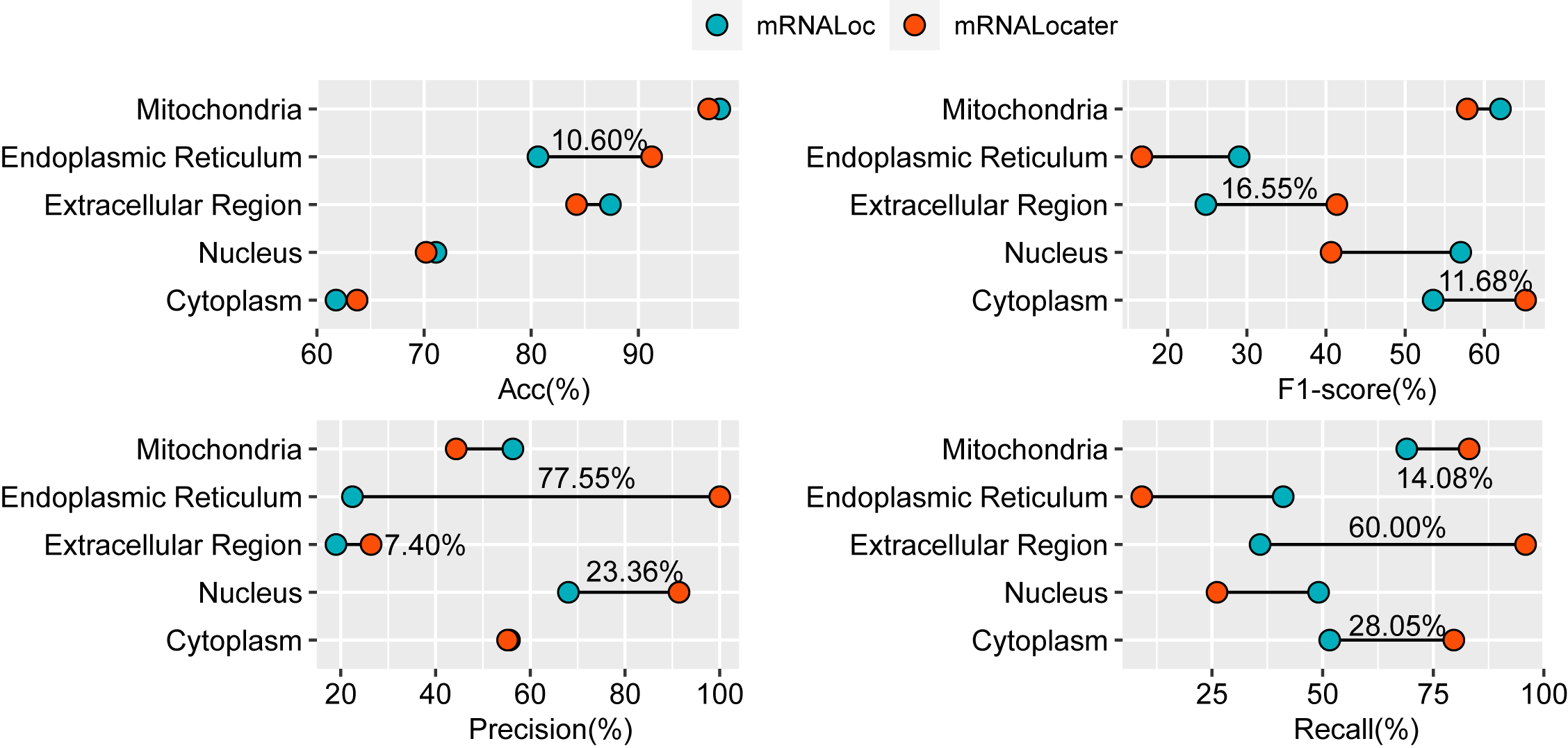
In order to better demonstrate the predictive accuracy and generalization ability of mRNALocater, we compared it with the best mRNA subcellular localization predictor mRNALoc. The comparison was performed based on the same independent test datasets. our proposed method can respectively achieve the optimal Acc of 63.73%, 91.24%, 84.23%, 96.56% and 70.19% for predicting the metiond five subcellular locations, which is superior to that of mRNALoc. These results demonstrate the promisong performance and stability of mRNALocater.

In the result page, we provide the predictive scores of the five subcellular location for each sample, which is a good reference for users. And users can easily download the result data of their select.